Earlier this month, our Principal Application Scientists, Stuart Firth-Clark, and Nathan Kidley led an exclusive online webinar, focused on guiding attendees through the new structure-based functionality (such as GCNCMC in dynamics/FEP; ensemble covalent docking) and ligand-based functionality (such as QSAR workflow enhancements and new QM features) now available within Flare V7.
For those unable to attend the webinar, or looking to catch up, this article serves to summarize the session and the full range of Flare functionality which was demonstrated. We have annotated the headings with the timings of each section of the webinar, the recording for which can be requested via this link.
Introducing Flare V7 [00:48]
Flare is a rapidly developing CADD software platform, which was first released back in 2017. Over recent years we’ve run bi-annual updates, in response to customer feedback, and to provide users with accelerated access to the very latest scientific methods. Components which have been newly added or enhanced within the version 7 updates include:
- Hit Expander
- Torsion Analysis
- Quantitative SAR
- Quantum Mechanics for ligands
- Docking and scoring
- Custom Force Field torsion parameters
- Free Energy Perturbation
- Molecular Dynamics
Certain features are of course most tailored and appealing to medicinal chemists, while others are very much targeted towards computational chemists. Flare’s modern, extensible, user-friendly interface makes the platform approachable and easy-to-use for a full variety of users, while the Flare Python API ensures that the most technically minded computational chemists can make full use of the platform, through customizable, scriptable, and deployable science.
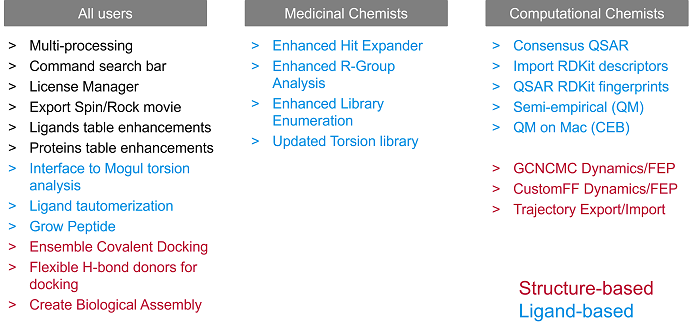
Figure 1: Newly added structure-based and ligand-based methods in Flare V7, categorized by their relevance to medicinal and computational chemists.
Introducing an example project: Caspase-3 [04:33]
To support in demonstrating the new features in Flare V7, we selected an example protein, Caspase-3. The caspase family of proteins are targets for inflammation and cancer, based on their involvement with apoptosis. The substrates for caspases are peptides, the enzyme hydrolyzing one of the backbone peptide bonds using an active site cysteine residue (Cys163 in Caspase-3).
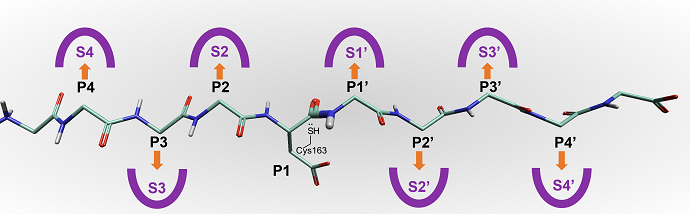
Figure 2: Representation to show a bound peptide in Caspase-3.
During the demonstration, we started with a crystal structure that has a covalently bound peptide, and explored, using Flare, how we can generate new ideas for peptide design, and find new molecules which are less peptidic in nature.
Downloading the protein into Flare and creating a biological assembly [07:18]
After downloading the crystal structure of Caspase-3, we discussed the application of one of the new features within Flare V7: the ability to create a biological assembly. When this feature is applied, Flare will automatically create either a dimer or tetramer of the chosen protein.
The new calculations panel in Flare V7 [08:57]
As soon as the ‘Start’ button is clicked within Flare, the ‘Calculations’ panel will appear. This new feature enables all calculations to be tracked and monitored, throughout the duration of the Flare session. For each calculation, once complete, users can access the log file and view the output from the chosen job.
Figure 3: The new ‘Calculations’ panel (right) displayed alongside the Caspase-3 protein in Flare.
Designing new peptides in Flare V7 [11:27]
After selecting the ‘E-chain’ of the prepared protein, we converted this into a ligand, enabling this portion to be the focus of the project. After a quick demonstration of using the new ‘Search’ box in Flare, we moved on to begin our project, using the new ‘Grow Peptide’ feature within the ligand-editing component of Flare. With this feature, new peptides can be built, simply by inputting the amino acid sequence into the dialog box.
A series of new peptides were created and moved to a new ‘Designs’ role within the ligands table.
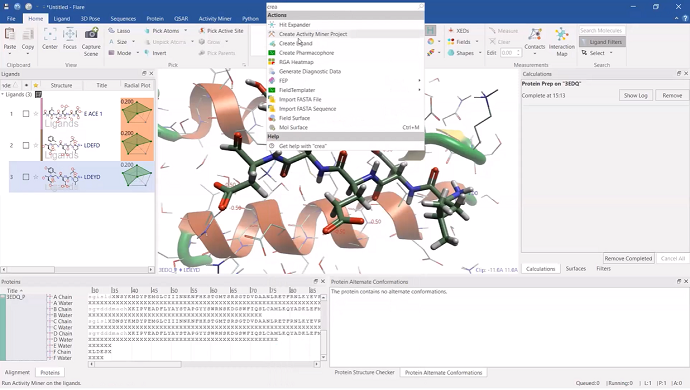
Figure 4: an example of a new peptide ligand being grown within Flare, with the ‘Search’ box overlaid.
Protein preparation [18:22]
After importing three new crystal structures of Caspase-3: 2CJX, 4PRY, and 5IAB, we explained how the new ‘Roles’ function in the proteins table works within Flare V7. The three new proteins were assigned to the ‘Unprepared’ role before the protein preparation process commenced.
All proteins were subsequently aligned and superposed to the originally prepared protein. A series of preparation actions were completed including the charging of picked atoms for pH7. During this section, we also showed another new feature in Flare V7 – the ability to run multiple jobs at the same time, without having to wait for jobs to finish before submitting the next task.
Ensemble covalent docking [26:20]
The three peptide ligand designs were selected and docked using the new ‘Ensemble Covalent’ docking feature in Flare. This new method enables users to consider protein active site flexibility, by using alternative active site conformations of the same protein when docking covalently bound inhibitors.
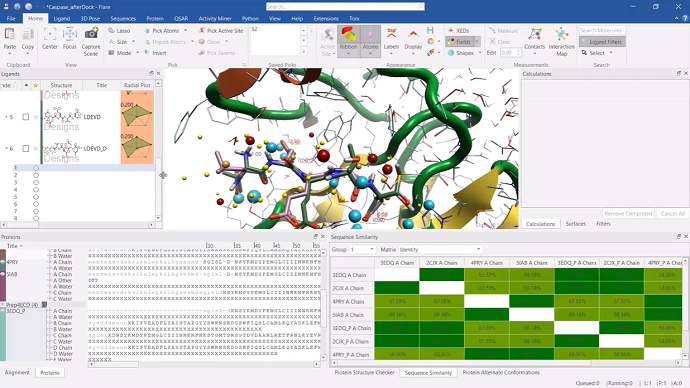
Figure 5: results from the ‘Ensemble Covalent’ docking experiment displayed in Flare.
Using the docking results for virtual screening in Blaze™ [33:10]
After running the docking simulation, Flare’s connection with ligand-based virtual screening server, Blaze, allowed us to download results from a virtual screening study that had been executed prior to the webinar. From this study, 250 commercially available compounds with similar electrostatics and shape properties to the reference peptide were downloaded into Flare, for further analysis.
The downloaded virtual hits are displayed within the ligands table, along with a large range of attributes. Thanks to the new ‘Pin Columns’ functionality, introduced within Flare V7, the user can select certain metrics to permanently appear as ‘frozen columns’ allowing the user to scroll across the Ligands table, while retaining the visibility of these pinned columns. We subsequently showed another new feature in Flare V7: the ability to calculate RDKit descriptors and demonstrated how these can be used with the ‘Filter’ function to identify ligands with desired features, for example, hiding compounds that had more than the specified number of amide bonds. Finally, we showed another new feature in the ligand preparation tool: the ability to enumerate tautomers and showed for one virtual hit that the tautomer can be easily changed.
Summary and concluding remarks [43:20]
Once the demo had concluded, Stuart finished the session with a quick summary of the areas that were explored within the webinar. These included:
- A range of new features found within the Flare V7 GUI
- Alternative workflows that are possible thanks to the new features
- New features that provide a more efficient way of traversing through complex workflows
- The incorporation of Blaze virtual screening results in Flare, outlining how Flare’s new features help users to prioritize the output molecules
Please follow this link to request access to the full session recording. We’d very much recommend that you catch up and see a live demonstration of the discussed Flare features. For those interested in exploring the platform firsthand, you can request a free evaluation, or download our free version of Flare, Flare Visualizer. We also offer a variety of flexible licensing options for students and academics.