Abstract
The correct assignment of the tautomeric and protonation states of ligands is critical for successful molecular modeling experiments in both ligand-based and structure-based design approaches, as it may have a significant effect on the shape and electrostatic properties of the ligands. Flare™, Cresset’s comprehensive computational chemistry software tool, features a ligand preparation module to automatically process ligands. However, there are of course occasions when you may want to further fine-tune the tautomeric or protonation state for specific ligands, based on your knowledge of the chemical series under study. In this article, we will demonstrate how this can be done using the ‘Library Enumeration' module within Flare.
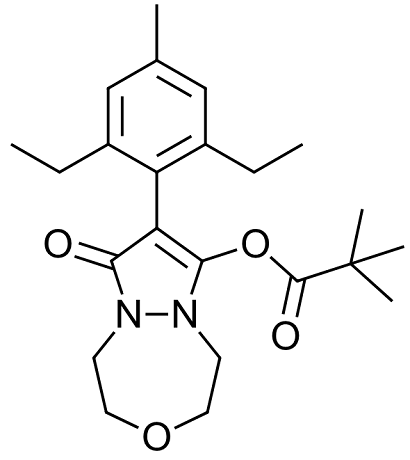
Figure 1. Structure of Pinoxaden.
Preparing the ligand
Pinoxaden (figure 1) is a post-emergence systemic herbicide that is used to control grass weeds. It does this by inhibiting Acetyl CoA Carboxylase (ACCase) binding in the carboxyltransferase (CT) pocket, which inhibits fatty acid synthesis. Pinoxaden is a prodrug herbicide, and the biologically active form is generated by the hydrolysis of the pivalate ester group, generating Pinoxaden acid with a pKa of 3.81.
Analyzing the crystal structure of Pinoxaden acid bound to the CT pocket of ACCase (PDB: 3PGQ), we can observe the bioactive conformation in the keto-enol form (Figure 2). The enol group is deprotonated due to its low pKa: the negatively charged oxygen occupies the oxyanion pocket of the CT pocket, maximizing electrostatic complementarity and making hydrogen bonds with the backbone amide of isoleucine 1735 and alanine 1627.
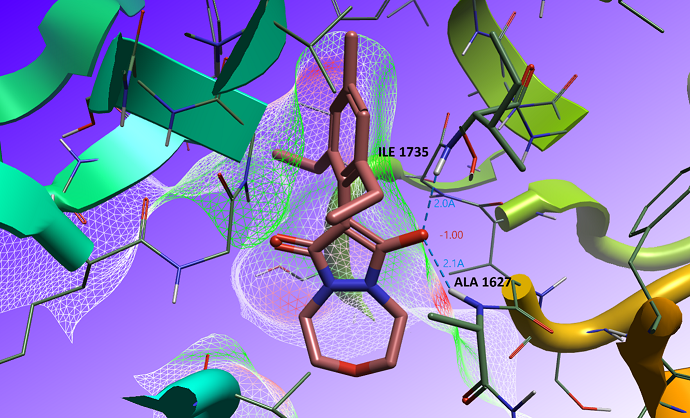
Figure 2: Bioactive conformation in the binding site of ACCase (PDB code: 3PGQ). The electrostatic complementarity (EC) surface is displayed and the coordinating amino acid residues within the oxyanion pocket are labeled.
However, when designing novel analogs of Pinoxaden, chemists could draw the pyrazolidine scaffold in either the dione or keto-enol forms, as shown in Figure 3. To prepare such ligands for docking, we would want all the ligands to be in the keto-enol tautomeric form and ideally deprotonated (bottom-right form in Figure 4). This can be done by manually editing the ligands within Flare: however, it is a time-consuming and tedious task in situations where many ligands need to be prepared.
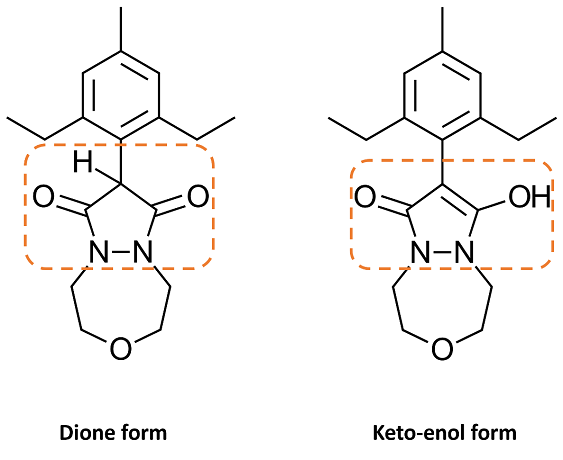
Figure 3: 2D representation of the dione and keto-enol forms of Pinoxaden.
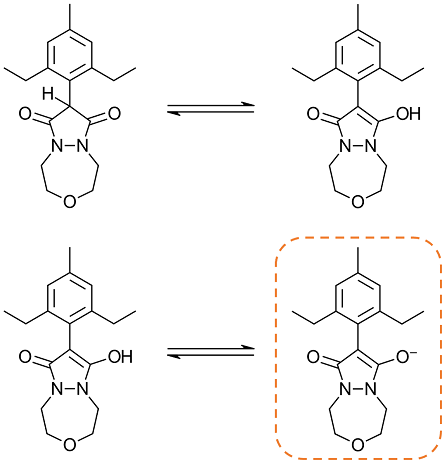
Figure 4: The tautomerization (top) and deprotonation reactions of the pyrazoline scaffold of Pinoxaden.
Utilizing Flare’s ligand preparation tool
As an alternative, we could use the ligand preparation module in Flare, which includes options for generating tautomers, as well as charging ligands appropriately for pH 7 (Figure 5). This workflow effectively transforms the dione into the protonated keto-enol.
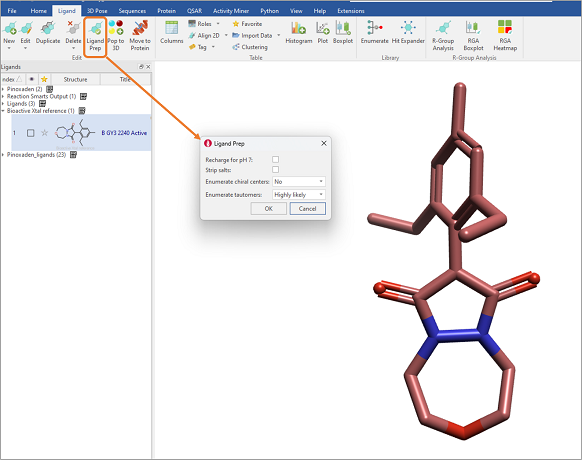
Figure 5: The Ligand Prep feature in Flare V7 has the functionality to enumerate tautomers, in addition to enumerating chiral centers, stripping salts, and re-charging for pH 7.
However, as this workflow uses a rule-based algorithm for protonating ligands, in this case the enol is not recognized as acidic, as the 1,3-dione is a less common substructure in life sciences chemistry, generating the top-right structure in Figure 4.
Fast generation of in silico chemical libraries and arrays
For this special case, we will use the Library Enumeration tool within the Flare to perform chemical modifications to all the ligands at once. The Library Enumeration tool in Flare is based on the RDKit enumeration method2, in which a Reaction SMARTS detailing the chemical transformation can be specified by the user to perform the desired chemical change. In our example, we are interested in making the chemical modification to the pyrazoline core only, meaning our Reaction SMARTS string need only encompass the matching SMARTS pattern for this functionality. Starting from the neutral keto-enol form obtained from the automatic ligand preparation workflow, Library Enumeration can transform the ligands directly into the enolate bioactive form we wish to use for docking, as shown in Figure 6.
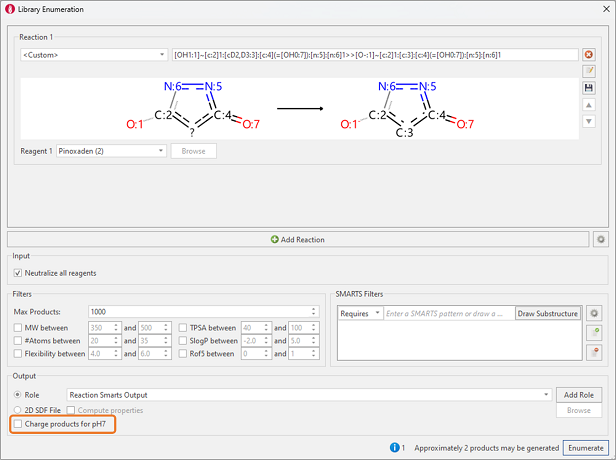
Figure 6. Library Enumeration is used to transform the neutral keto-enol form into the deprotonated keto-enol bioactive form.
Furthermore, if the Pinoxaden analogs are drawn in the dione form (left in Figure 3) we could completely skip the ligand preparation step and use Library Enumeration to transform the 1,3-dione into the desired deprotonated keto-enol form using the reaction shown below (Figure 7).
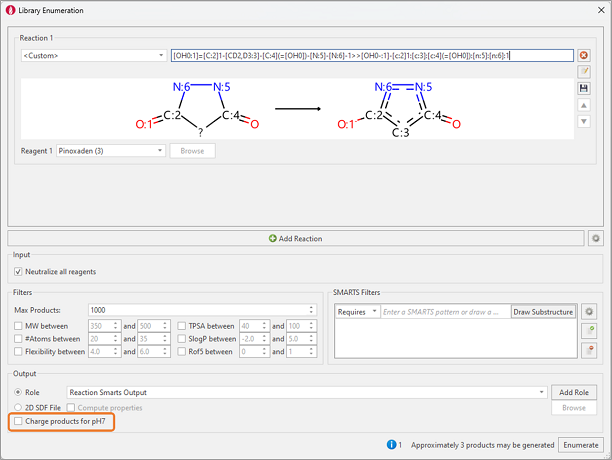
Figure 7: Library Enumeration is used to transform the pyrazolidine-1,3-dione form into the deprotonated keto-enol bioactive form.
In the above reactions, to ensure that the product is in the desired deprotonated form, it is important to keep the ‘Charge products for pH7’ options unselected to ensure the results are in the desired acidic form.
We have now avoided the need to manually edit our ligand designs one by one and, instead, can efficiently prepare the ligands in the desired form in Flare.
Conclusions
This example demonstrated how the enumeration tool can be used to change the tautomeric and protonation state of a ligand. In addition to the option of using custom reactions, the Library Enumeration tool in Flare supports more than 50 common synthetic chemical reactions. These can be used ‘out of the box’, to support the generation of in-silico ligand libraries.
Request a free evaluation of Flare today to further explore its full portfolio of molecular modeling capabilities.
References
- Muehlebach M, Boeger M, Cederbaum F, Cornes D, Friedmann AA, Glock J, Niderman T, Stoller A, Wagner T. Aryldiones incorporating a [1,4,5]oxadiazepane ring. Part I: Discovery of the novel cereal herbicide pinoxaden. Bioorg Med Chem. 2009 Jun 15;17(12):4241-56. doi: 10.1016/j.bmc.2008.12.062. Epub 2009 Jan 3. PMID: 19167895.
- https://www.rdkit.org/docs/RDKit_Book.html#chemical-reaction-handling


