Understanding ligand-protein interactions through visualization is an essential part of scientific reasoning and is a crucial decision-making process that computational and medicinal chemists routinely use. Therefore, scientists need handy tools that can quickly and efficiently perform both simple and complex selections of atoms or residues to highlight the critical information in protein-ligand complexes. Such tools can also play an important part in helping the preparation of the molecular system, for example, correctly setting up protonation states of residues before docking and molecular dynamics simulations. Efficient selection tools can therefore reduce the analysis time and help to deliver the project results within the required timeline. Furthermore, selection tools are indispensable for creating compelling and informative images for project reports or scientific publications.
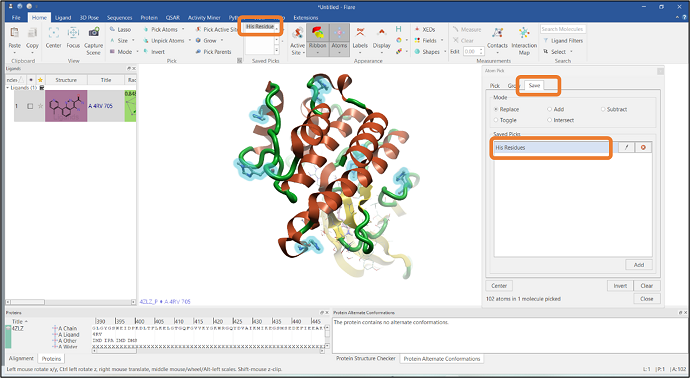
Figure 1: The Atom Pick feature can be accessed through the Home tab in Flare.
Flare™ is the comprehensive software platform for structure and ligand-based molecule design. The Flare GUI includes many practical functions for selecting atoms and residues of interest, including the Atom Pick panel which can be used to define more complex atom or residue selections. The "Atom Pick" panel is available by clicking the arrow button found in the "Home" tab (See Figure 1).
Examples of complex atom selections in Flare
Select active site residues
With one or more ligands and proteins selected, clicking on the "Active Site" button will pick the residues in the protein's active site which have at least one atom within the specified radius of the ligand (See Figure 2): this function is equivalent to the Pick Active Site button in the "Home" tab. In contrast, clicking the "Not Active Site" button will pick the atoms that are NOT part of the active site definition.
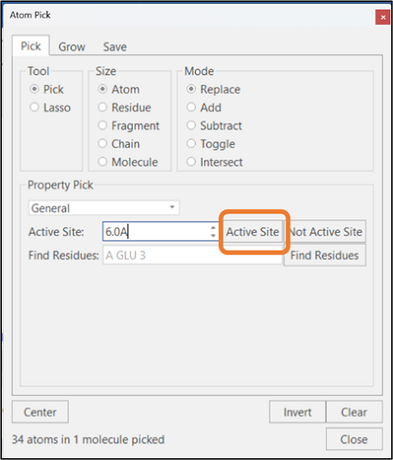
Figure 2: Selection of active site residues.
Select a particular amino acid residue (e.g., His) found in the protein sequence
It is often important to check the tautomeric/protonation state of Histidine residues as both docking and molecular dynamics simulations require the correct tautomeric/protonation state of these residues.
You can easily select a particular residue from the Property Pick menu: select the "Residue" option and click the HIS button to pick all the Histidine residues in the protein sequence (Figure 3).
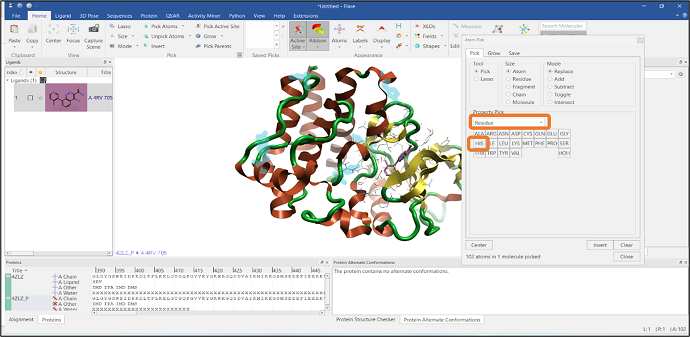
Figure 3: Selection of all Histidine residues using the Atom Pick tool.
It is also possible to save the pick for future reference by going to the "Save" tab in the "Atom Pick" panel. In this example, the current selection has been saved as "His Residues" which will subsequently appear in the "Saved Picks" list in the "Home" tab (see Figure 4).
The immediate environment around histidine residues can determine the protonation/tautomeric state of those amino acid residues. It is therefore often important to highlight the neighboring environment of a particular amino acid. This can be done easily using the "Grow" and "Pick Parents" buttons located on the ribbon of the "Home" tab (Figure 5).
Note: It is possible to change a Histidine residue's tautomeric/protonation state by clicking the "Change Tautomer" and "Change Charge State" buttons found in the "Protein" tab.
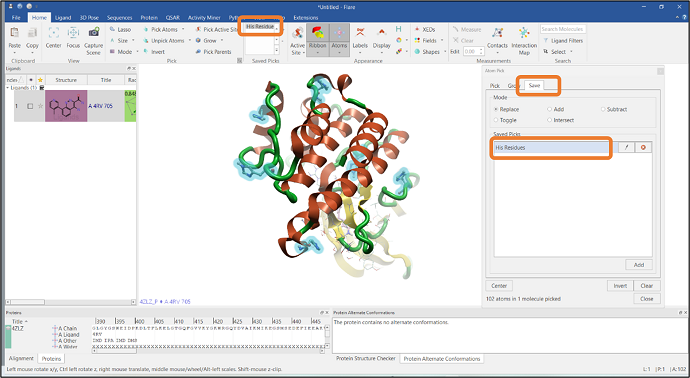
Figure 4: Saving the Histidine residue pick.
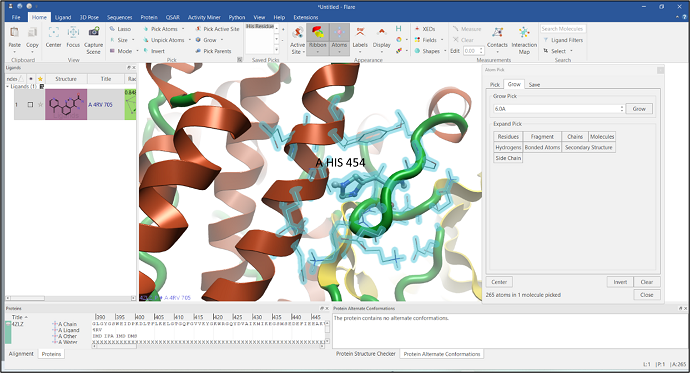
Figure 5: Grow the surrounding environment of a selected residue.
Select water molecules within the active site
When analyzing the results of a molecular dynamics simulation, we often want to highlight how the water molecules are interacting with the ligand at a particular time in the simulation trajectory. One way to pick these water molecules in Flare’s "Atom Pick" panel is to type "HOH" in the "Find Residue" text box and then click on the "Find Residues" button (see Figure 6). In the next step, click the "Subtract" option from the "Mode" section and click the "Not Active Site" button. This will subtract all the water molecules that are not part of the active site (see Figure 6).
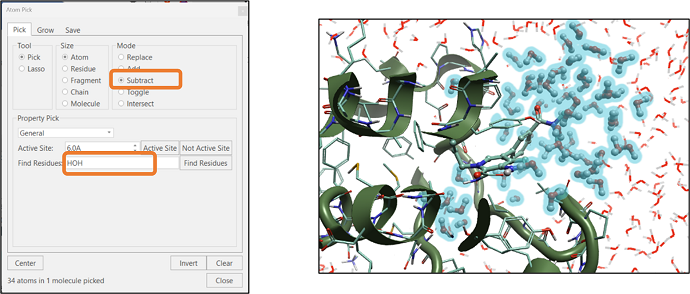
Figure 6: Selection of active site water molecules using Atom Selection tool.
Selecting protein side chain atoms
Protein minimization is an essential step prior to running molecular dynamics simulations to remove clashes between atoms. The "Protein Structure" option in the "Property Pick" pulldown gives access to different options for atom selection. The option "Side Chains" can be selected (see Figure 7) after which a minimization on these selected side chain atoms can then be performed.
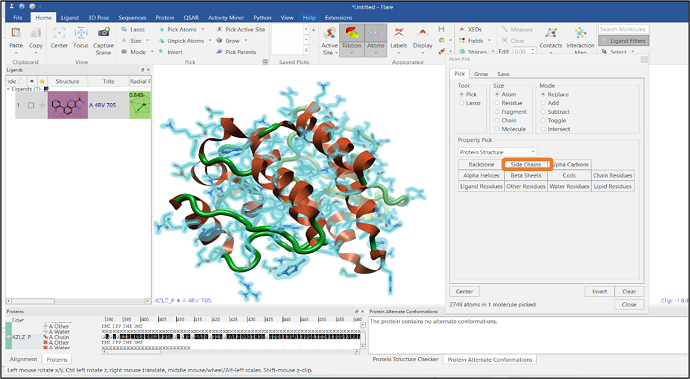
Figure 7: Selection of side chain atoms of a protein.
Selection of residues at the interface of a protein-protein complex
It is often required to visualize the residues at the interface of a protein-protein complex and see how these interact with the other chain. In this example, we have imported a PDB file that has two chains: chain A and chain E, represented in gray and green colors respectively (Figure 8). We will pick the residues of chain E that are in contact with chain A using Flare’s Atom Pick tool. In the first step, atoms in chain A are picked by typing "A" in the "Find Residues" box (see Figure 8). Residues that are within 6Å from the atoms of chain A are then added to the pick by pressing the "Grow" button in the "Grow" tab and expanding the pick using the "Residue" button (Figure 9). Finally, chain A atoms are removed from the pick by using the "Subtract" mode from the Mode section (see Figure 10). Alternatively, you can select chain A residues that are in contact with chain E
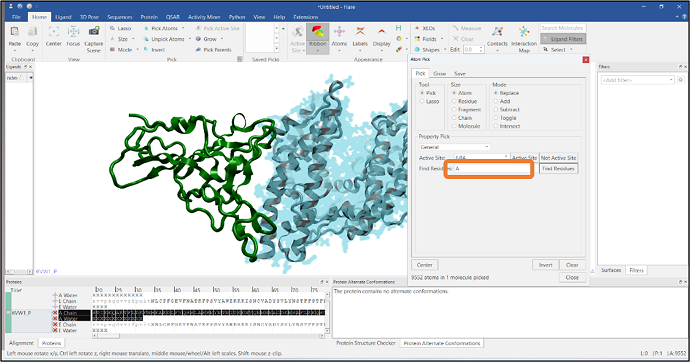
Figure 8: Selection of all the atoms from chain A.
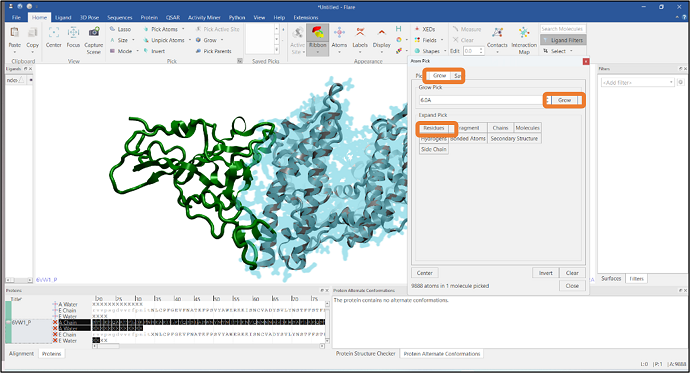
Figure 9: Selection of residues that are within 6Å from the atoms of chain A.
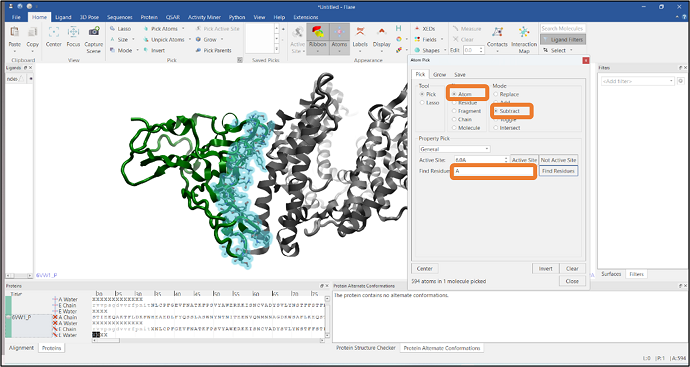
Figure 10: Selection of interfacial residues by subtracting the atoms of chain A.
The above examples are just a few atom/residue selection methods that can be easily performed using the Atom Pick feature in Flare. Besides the GUI, the Atom Pick options can be accessed through Pyflare, a dedicated python binary where all the complex selections can be done through python commands.
Deliver the project results you need by running complex atom selections in Flare
In summary, the atom/residue selection panel enables researchers to deep dive into the protein-ligand system to understand the various processes in a much more intuitive way, and the Flare Atom Pick method provides a faster and easier way to access all the complex selection options.
Request an evaluation of Flare to explore complex molecular models with ease.