Growing up as a child, I thought that antibiotics were the magical answer for most medical issues, be that a persistent sore throat or a high fever. Little did I know that antibiotics were only effective if my illness was only caused by bacteria, or that the rate these drugs were (and continue to be) prescribed has been a leading contributor to Antimicrobial Resistance (AMR) – the ability of pathogenic microorganisms such as bacteria and fungi to become resistant to and defeat the drugs designed to kill them.
With the urgent need to fight AMR, clinicians are often placed in the difficult position of having to quickly decide when to prescribe antibiotics, a complex decision which involves weighing the patients’ health against the potential collateral harm to society and healthcare at large. Diagnostics can contribute towards fighting AMR by providing rapid, highly accurate and reliable information to support a clinician’s decision making and pave the way towards personalised prescribing for antimicrobials: using the narrowest-spectrum agent that the pathogen is susceptible to, at the right dose, for the shortest duration.
Despite numerous diagnostic products helping to fight the overprescription of antimicrobials, as of yet none of them seem to provide a clear-cut answer quickly enough to enable clinicians to optimise prescribing. There is clearly a need for more effective diagnostics to help tackle AMR. The question is, what do clinicians need and how do their requirements align with the capabilities of key diagnostic methods in the field?
The antimicrobial resistance crisis
In 2020, the European Centre for Disease Prevention and Control (ECDC) reported that there was more than a three-fold difference in antibiotics consumption between EU countries. That same year my home country, Cyprus, was number one for antibiotic consumption in Europe, which was heart-breaking for me. If antimicrobials lose their effectiveness, this means that we lose the ability to treat infectious diseases and infections. In 2019, it was estimated by the CDC that AMR was responsible for at least 1.27 million deaths worldwide, while it has also been estimated that the continued rise in resistance will result in 10 million deaths annually by 2050.
The more microorganisms are exposed to antimicrobials, the better they become at surviving them. A prudent and selective use of antimicrobials is therefore essential to maximise their therapeutic effect, whilst minimising the development of AMR. Despite this, according to the CDC, about a third of antibiotic use in people is neither needed nor appropriate. One reason for this is a lack of suitable, widely adopted rapid diagnostics, which help provide clinicians with the information needed to make treatment decisions.
Tackling AMR: diagnostic requirements
The suitability of a diagnostic can broadly be determined by considering the following key characteristics: information, speed, cost and usability.
Providing information to clinicians
Diagnostics can contribute towards fighting AMR by providing highly accurate and reliable information on what is causing an infection, how best to kill it and how the patient is responding.
- What is causing it? The treatment of infectious diseases strongly depends on the pathogenic microorganism causing it. However, bacteria and viruses can often both cause similar symptoms. This means it is often impossible to tell whether someone has a viral or bacterial infection from their symptoms alone. Diagnostics that can rule out bacterial infections can thus help avoid the unnecessary use of antibiotics.
- How to kill it? Diagnostics that can specifically identify which microorganism is causing the infection, as well as its resistance profile against specific antimicrobials, would also help to determine the right antimicrobial to use at the right dosing.
- How is the patient responding? Diagnostics that show how the infection is manifesting on the patient would help to weigh patient risk and whether antimicrobials are suitable. Identifying a potentially pathogenic organism in a patient may not mean it is responsible for the patient’s condition. In the cases where it is, the patient could still resolve the infection without antimicrobials by relying solely on their own immune system.
Speed of diagnosis
The undisputed requirement for AMR diagnostics to become widely adopted in clinical settings is that they need to be fast. Diagnostic information returned from within a few minutes to a maximum of a few hours would help to cover the needs of both the fast pace of primary care (10-minute GP appointments) and the urgency of acute and emergency care, were mortality increases with each hour (such as sepsis and HAP/VAP cases). With fast diagnostic turnouts, occupancy rates and the number of medical appointments at hospitals can be reduced.
Cost and usability
The question of how to tackle antimicrobial resistance is a global challenge that needs to be addressed on a global scale. Any diagnostic solutions therefore need to be affordable, to make them accessible in all locations. It is often suggested that for a test to be feasible, it must cost less than a course of antibiotics.
From a usability perspective, diagnostics should also be universally easy and safe to use, at the point-of-care and at the bedside, with minimal training and no lab infrastructure required. Results should be accessible and easy to interpret, in the fashion of sample-to-answer, and easy to communicate to support patient dialogue. Having such a diagnostic could allow potentially for nurses and pharmacists to also diagnose and treat patients with infectious diseases, hence relieving the pressure on primary care.
Diagnostic methods for AMR
The common diagnostic techniques for identifying pathogens and profiling their antimicrobial resistance are largely classified as ‘phenotypic’ or ‘molecular’. Phenotypic methods are based on how microorganisms appear (e.g. size and shape) or how they manifest their presence (e.g. growing or producing proteins), which is used for identification. These methods are also based on how the microorganisms respond to the presence of antimicrobials, which are used for antimicrobial susceptibility testing. Meanwhile, molecular methods are based on recognising the microorganism’s distinctive genetic profile.
Phenotypic methods
Phenotypic methods can provide information on the “what is it” and “how to kill it” questions and are powerful as they provide a direct indication of the susceptibility of a given microorganism to an antimicrobial agent, at defined concentrations. Some of the key methods include culture-based methods and protein detection.
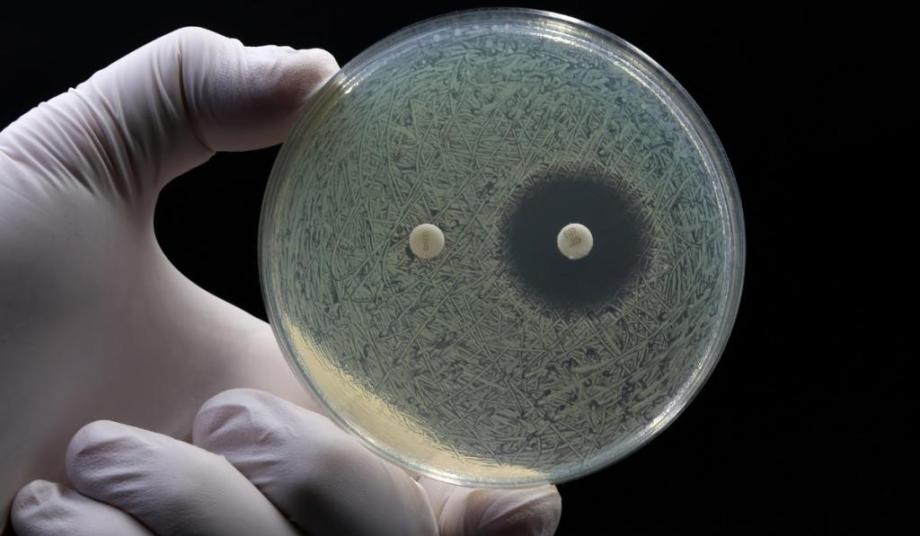
Culture-based methods
Culture-based methods are the gold standard for determining bacterial infection and antibiotic susceptibility. This usually involves subjecting a patient sample to favourable conditions for growth. A positive bacterial infection is then confirmed when colonies of bacteria appear after a few days.
Growth of bacteria can also be carried out in the presence of different types of antibiotics to evaluate their resistance profile (known as antimicrobial susceptibility testing, or ‘AST’) and determine the right type and dosing. Specific species can be identified by using selective media for growth and species-dependent staining techniques, or by using methods based on the microorganism’s enzymatic activity.
The main limitation of culture-based methods is speed, requiring an initial culture step for 24–72 hours and even longer to confirm antimicrobial susceptibilities. In most cases, this occurs well after the initial prescription decision has been made.
Protein detection methods
Protein detection is another phenotypic technique, where specific proteins associated with specific viruses or bacteria are detected. These methods can be used for both the detection of a bacterial antigen and also for serology – i.e the detection of antibodies against a viral infection. Many protein detection based diagnostics are based on the antigen-antibody interaction, such as the ELISA method (Enzyme-Linked ImmunoSorbent Assays). In an ELISA, the antigen or antibodies of interest from the input sample are captured by a corresponding antibody or antigen immobilised on a microtiter plate. A secondary antibody with a fluorescent label is introduced to detect the antigen/antibody complex.
Whilst protein detection is a good technique for pathogen identification it does not provide information as to which antimicrobial would be most appropriate. Lateral flow tests (LFTs) are a popular example of a phenotypic method based on the detection of specific proteins. Several LFTs are currently available for identifying infectious viruses, such as A/B Influenza, Sars-CoV-2, HIV, Hepatitis, Ebola and Zika. The identification of AMR strains is also possible using LFTs by detecting proteins that are known to be associated with AMR.
What’s more, LFTs meet all the prerequisites defined by the World Health Organisation for an ideal point of care test, being affordable, sensitive, specific, user friendly, rapid, robust, equipment free and deliverable. The main constraint of LFTs is that they typically only allow the detection of a single pathogen at a time.
Molecular methods
Molecular methods are based on the detection of genetic material in a sample. These methods can detect infectious pathogens and their associated drug resistance directly from a patient’s sample, by detecting the microorganism’s DNA and genes associated with antimicrobial resistance. Therefore, the method relies on having a comprehensive database of these genes and their antimicrobial resistance and cannot detect any resistance due to new, uncharacterised mechanisms. Even in the case where a genetic marker is identified, the method cannot distinguish whether it will be active or not to confirm that resistance will take place phenotypically.
Polymerase Chain Reaction (PCR) and DNA Microarrays
Molecular based tests developed to detect certain infectious diseases are called molecular based assays. The most common molecular assays are based on the Polymerase Chain Reaction (PCR). When a specific segment of DNA of interest (e.g. part of a virus’ DNA) is present in the sample it gets amplified to return a measurable signal to confirm its presence. DNA-Microarrays is a similar method where a microarray chip is used to simultaneously detect different specific pathogens and vast numbers of different resistance genes in a single test.
A big advantage of molecular based assays is that they are generally faster and more sensitive than culture-based methods, returning results within a few hours. In contrast to LFTs they are easy to multiplex, allowing screening for tens of pathogens/genes in a single test. They are, however, more expensive than both the other methods and require specialised equipment and trained staff to operate them, making them unavailable at the point of prescribing. Compact, user-friendly lab-on-a-bench-like systems such as the VERIGENE System (Luminex), the BioFire System (Biomerieux) and T2Dx System (T2 Biosystems) have the potential to enable clinicians to rapidly identify pathogens directly from a patient sample, all within two hours.
Whilst molecular assays are excellent for answering the “what is it?” question, the method does not distinguish infection from asymptomatic colonisation, making the interpretation of results a challenge particularly when many pathogens are detected at once.
Genome sequencing and metagenomics
Whereas conventional molecular diagnostics can identify tens of common pathogens and resistant markers at a time, metagenomics allows the identification of the entire community of microorganism DNA within a sample (microbiome) using methods common to whole genome sequencing (WGS).
In WGS, the whole genome of an organism is determined in one process, by detecting in a sequential order each nucleotide (the basic building block of DNA) present in a sample. The derived sequences are then checked against known databases of unique pathogen sequences and genetic resistance markers, to identify the microorganism and its antimicrobial resistance. Metagenomics offers the capacity to detect all potential pathogens —bacteria, viruses, fungi and parasites — and all genes involved in AMR in a single sample. It therefore has the potential to be a great clinical diagnostic tool as well as a research tool for unraveling new pathogens and emerging resistance factors.
Another powerful capability of metagenomics is the capacity to study human host immune responses to infection (transcriptomics) making it possible to answer all three key diagnostic questions in one test: “what is it?”, “how can we kill it?” and “how is the patient doing?”.
Despite these advantages, metagenomics is not routinely performed in clinical practice, as turn-around times for sequencing are too long (the sequencing run alone takes >18 hours). It also requires specialised labs and staff, has a higher cost compared to all the other methods and has the same limitations as molecular based assays.
Genomics providers are, however, increasingly expanding their infectious diseases portfolio, aiming towards making their solutions more comprehensive, portable, cheaper, and faster.
For example, Illumina has established a collaboration with IDbyDNA to deliver sample-to-answer solutions, which have been used by Synergy Laboratories to offer the first comprehensive NGS-based test for urinary tract infections (UTI). The test includes ~100 uropathogens and 371 select genetic markers of antibiotic resistance.
Oxford Nanopore Technologies has also recently announced collaborations with BioMérieux and leading China-based diagnostic companies, towards the development of in-vitro diagnostic (IVD) tests for metagenomics and infectious disease.
Biomarker testing
Another method, beyond the traditional phenotypic and molecular pathogen detection methods but still relevant for infectious diseases and AMR diagnostics, is biomarker testing. Biomarkers are biological molecules found in blood or other body fluids that are associated with a condition or a disease – and measuring them is a way of characterising the patient’s response to the disease. The two most common biomarkers relevant to infectious diseases are C-reactive protein (CRP) and procalcitonin (PCT).
CRP/PCT tests are often based on the ELISA method, mentioned previously. The level of CRP and PCT biomarkers in the blood increases during an infection and is often higher during a bacterial infection than a viral one. The level of biomarkers and speed at which they are being produced can be used to differentiate bacterial from most viral infections. However, this is not always the case. Some viruses (such as COVID-19) or autoimmune diseases can result in a similar biomarker increase as bacterial infections, making the interpretation of these tests very challenging. As a result, biomarker testing has been more useful as a complementary test , interpreted alongside clinical symptoms and the experience of healthcare professionals rather than a standalone sample-to-answer diagnostic.
Emerging diagnostics for AMR
Due to the urgency of the AMR challenge, great efforts are being made in innovating and reinventing traditional methods, to push towards affordable, accurate, fast and easy-to-use tests for identification and susceptibility testing. There are a number of nationally funded programs promoting the development of new tests. For example, the National Institute of Health has led the Antimicrobial Resistance Diagnostic Challenge which awarded Visby Medical’s single-use, instrument-free rapid PCR test $19 million in 2020. The Longitude Prize is also a similar competition underway, offering an £8 million prize led by Challenge Works in collaboration with Innovate UK.
There are many solutions currently being developed or recently released to market. Some of the most notable include:
Next generation single cell culture-free spectroscopy
Next generation single cell methods aim to reduce the days’ or even weeks’ long cultivation step by developing high-sensitivity techniques able to identify pathogens and pathogen growth at the single cell level (e.g. detecting a single bacterium multiplied to two or three bacteria rather than a whole colony, or measuring bacteria metabolic activity as a sign of growth).
Among these methodologies, single-cell spectroscopic techniques, specifically Raman spectroscopy and optical photothermal infrared spectroscopy (O-PTIR) are showing promise for rapid and accurate detection of pathogens and antibiotic resistance. Raman and infrared spectroscopic techniques identify pathogens by detecting the distinctive vibrations of the molecules comprising each pathogen – like a fingerprint. Recent advances in artificial intelligence methods have enabled the rapid and accurate translation of complex spectroscopic “fingerprints” measured to pathogen identification.
Beyond pathogen identification, these single-cell spectroscopic techniques have also demonstrated potential for measuring microbial resistance to antibiotics by monitoring metabolic activity in the presence of antibiotics.
The Nostics platform, leverages Raman spectroscopy, nanotechnology and AI, and aspires to enable testing to every corner of the world with a handheld device detecting bacteria and fungi in less than 15 minutes.
Single-cell technologies powered by droplet microfluidics or electrochemical impedance spectroscopy also offer great promise. Droplet microfluidics in combination with high efficacy fluorophores enable the detection of growth from single bacterial cells under antibiotic exposure within one hour. Electrochemical impedance spectroscopy is a technique that has been shown to successfully identify growth profiles and has been used to confirm antibiotic susceptibility in just 90 minutes using a 3D-printed biosensor.
Biomarker combination
As discussed earlier, as it stands there is no known single biomarker that can reliably differentiate the origin of an infection, however recent work suggests that analysing a variety of biomarker combinations (e.g. TRAIL, IP-10 and CRP) can tell bacterial from viral infections more accurately.
MeMed BV is an uplifting example, being the first FDA-cleared biomarker test that distinguishes between bacterial and viral infections in just 15 minutes. The MeMed Key system is an easy-to-use, compact benchtop platform that detects simultaneously up to four different biomarkers. It uses artificial intelligence methods to translate the complex signaling of the immune system into an actionable diagnostic outcome.
Instrument free PCR
Advancements in microfluidics, photonics and nanotechnology are partly responsible for the emergence of more miniaturised and cheaper diagnostic systems. An example is Visby Medical’s first of its kind instrument-free PCR test system that is able to diagnose for up to three pathogens per single test in under 30 minutes. Visby Medical’s test runs on a handheld single-use device which integrates a microfluidic reaction chip with integrated sophisticated micro-heaters for PCR amplification, and a microfluidic detection module.
As more portable, accurate, rapid tests such as the ones mentioned in this section become more widely available and adopted, they could be a powerful tool in the fight against AMR.

The ideal AMR diagnostics solution
There are no clear winners and losers in the methods available for improving diagnosis to tackle AMR. Each of the methods discussed offers different levels of information, often complementary to each other, and each comes with its own pros and cons when it comes to applying them in a clinical setting.
Culture-based methods remain the golden standard in terms of clinical relevance and are the most trusted by clinicians, but are too slow to win the fight against AMR. Molecular-based methods are the tool of choice for research purposes, with high accuracy and throughput, and with IVD systems emerging in hospitals, often as a support to conventional testing. Protein detection and biomarker testing methods are the current methods of choice when it comes to fast, cheap, accessible outcomes, however their results are of higher uncertainty and less clinically useful.
For a diagnostic in the field of AMR to have disruptive impact it would need to be widely adopted globally, in a similar manner to how bacteria cultures are adopted today. For that to happen it would have to be indisputably useful with a low adaptation barrier. In addition, a single point-of-care system carrying all the steps and generating all the information required for going from sample-to-answer would be more attractive than multiple systems offering complementary information (often systems with incompatible consumables and workflows).
Perhaps the ideal diagnostic solution lies at the intersection of all the different methods and multidisciplinary technological advancements mentioned above. Could the ideal diagnostic be designed through a single modular system that combines the strongest features of each method? This might entail a bio-instrument capable of molecular detection for pathogen identification, answering the ‘what is it?’ question, single-cell spectroscopy for antimicrobial susceptibility testing answering the ‘how to kill it?’ question, and biomarker testing for host-response characterisation to answer the ‘how is the patient doing?’ question. Perhaps then a sophisticated AI algorithm could weigh the outcomes from all three key questions to report a suggestion for personalised treatment.
A flexible system equipped with microfluidic capabilities, optical or electrical sensing and a collection of various accompanying reagents should in principle be capable of automatically conducting a test from any of the key methods. Such a system could leverage advancements in photonics to reduce detection times, as well as microfluidics and nanotechnology to reduce size, costs and increase throughput, and finally Artificial Intelligence to improve data interpretation.
No doubt building such a system would be a mountain to climb, but not unlike similar mountains scientists and engineers have conquered in the field of diagnostics in the past, and not unlike systems I enjoy dreaming of and am enthused to develop.